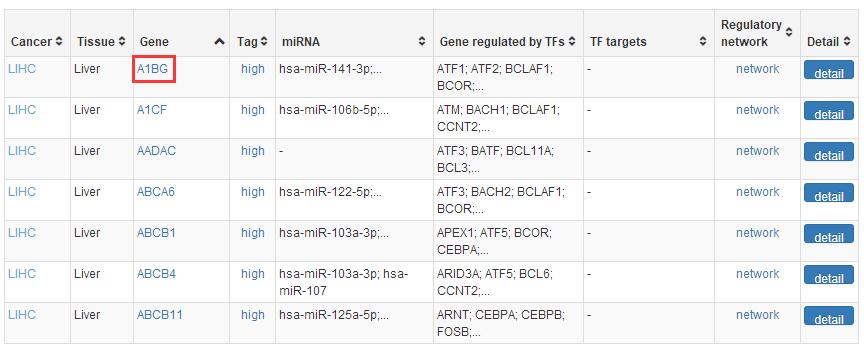
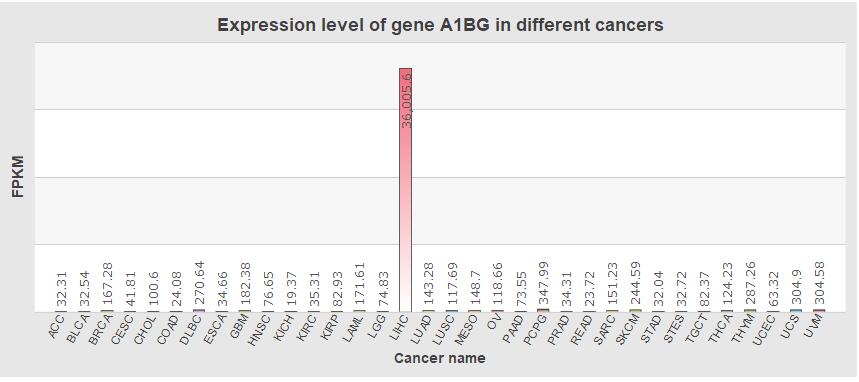
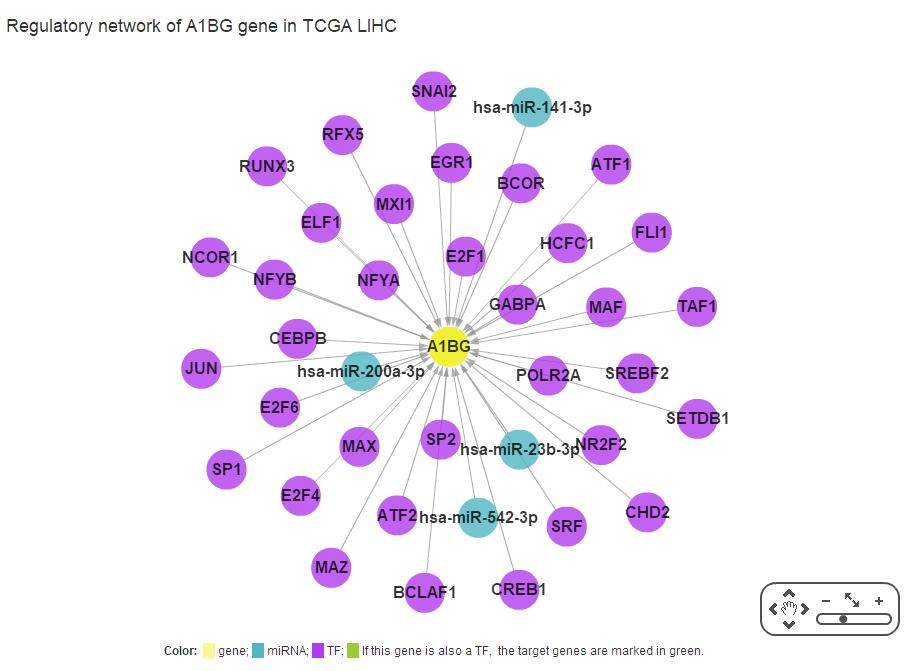
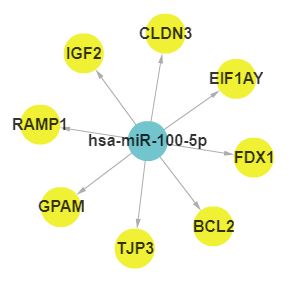
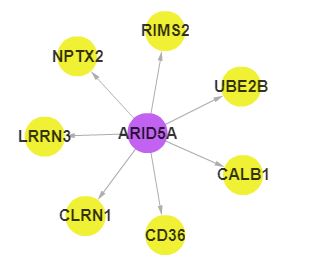
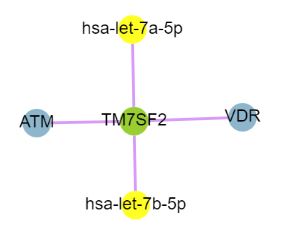
In SEGreg, SEGs is identified by SEGtool, which based on Fuzzy c-means, Jaccard index and greedy annealing method and outerperform the existing tools. Browse by tissues or genes as mentioned above, users will get the informations of SEGs presented in table format, in which there is a column named "Tag", by clicking the content in this column, users will see highly/lowly expressions of that gene in various cancer types/tissues. For example, gene A1BG highly expressed in LIHC, click the tag column of it,you will see expressions of it in different cancers, like the figure shown in below: